The use of in vivo synthetic biology has seen remarkable efforts in the greener production of chemicals and compounds at a commercial scale. However, such systems tend to be limited in their productivities. We are interested in developing yet more efficient platforms for biomanufacturing. L-malic acid, a compound with significant applications in food, pharmaceutical, and chemical industries, represents an excellent target for innovative biomanufacturing strategies.
In collaboration with Prof. Mattheos Koffas, we are developing an E. coli-based chassis for the cell-free production of L-malic acid (Figure 1). Using flux balance analysis, several metabolic engineering targets were identified to conserve carbon-flux in the biosynthetic pathway. These genes were categorized into essential and non-essential groups for depletion either via a knockout or a secondary elimination strategy. Mutant strains, thus generated, are being evaluated for the increased accumulation of malate in a cell-free reaction (Figure 2).
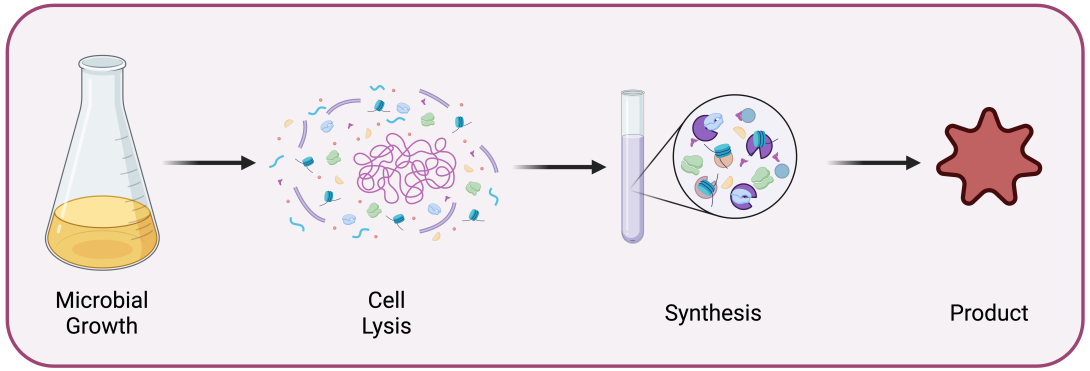
Figure 1. Schematic illustration of a cell-free system. The pathway of interest is produced using a microbial source during the growth phase. Subsequently, the cells are lysed and the crude extract thus obtained is used for the biosynthesis.
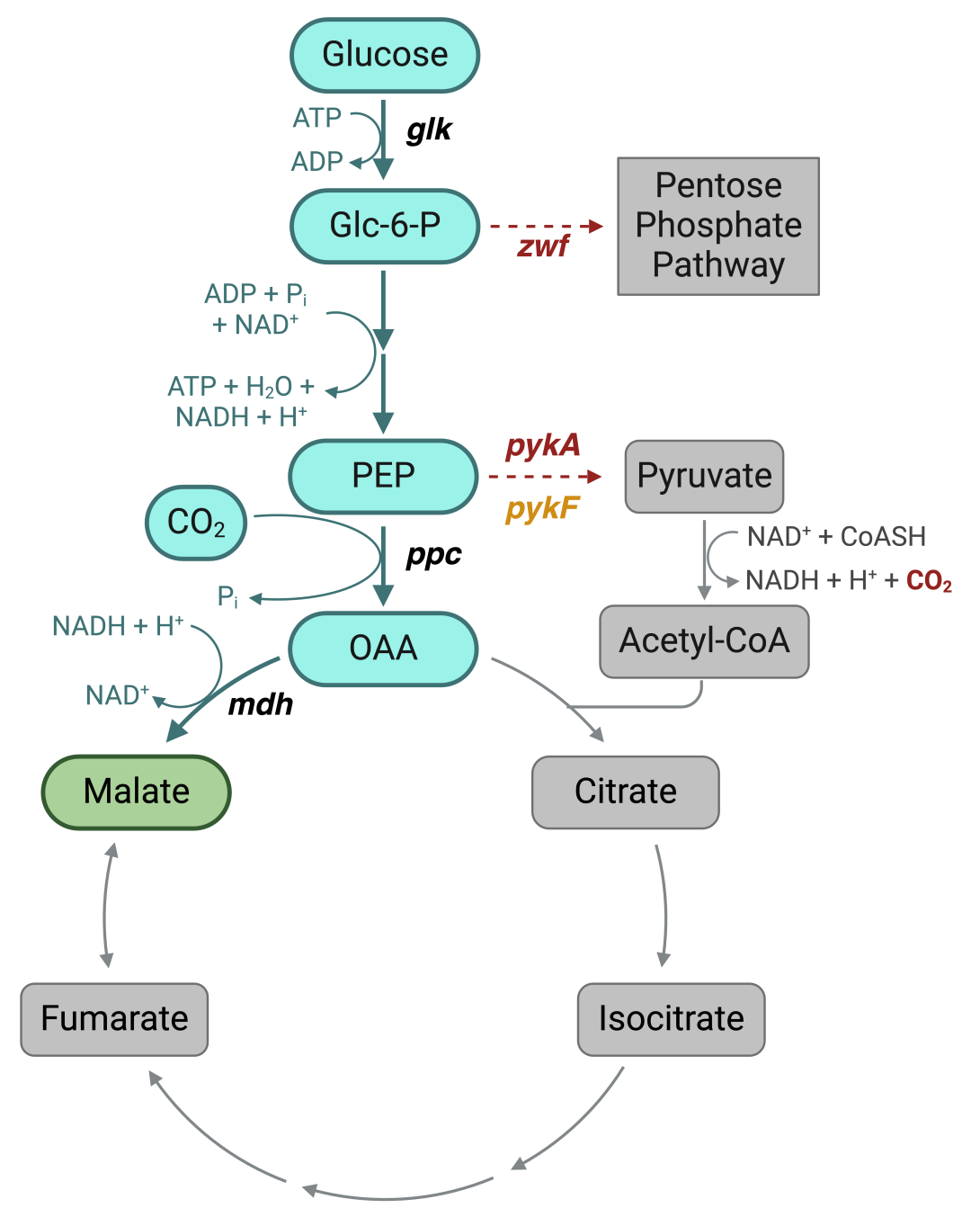
Figure 2. The engineered pathway for malate synthesis, using glucose as a carbon source in E. coli. The flux is enriched in the pathway in blue and depleted in the steps shown in gray. Additionally, a few targets for knockout have been shown in red and those for secondary elimination are in orange. glk: glucokinase, ppc: phosphoenolpyruvate carboxylase, mdh: malate dehydrogenase, zwf: glucose-6-phosphate dehydrogenase, pykA: pyruvate kinase II, and pykF: pyruvate kinase I.
