Perhydrolase-polydopamine composite coatings possessed potent antiviral activity, and dramatically reduced the infectivity of a SARS-CoV-2 pseudovirus within minutes [Wang et al. Sci. Rep. 11, 12410, 2021]. The single-step approach enables rapid and facile fabrication of enzyme-based disinfectant composite coatings with high activity and stability, which enables reuse following surface washing.
Rapid deactivation of virus particles on contaminated surfaces is critical to achieve reduced viral load and transmission. To this end, we examined two viruses, including a VSV-g pseudotyped lentivirus and a SARS-CoV-2 spike pseudotyped lentivirus, both carrying an Egfp reporter gene. Perhydrolase-polydopamine composite coatings possessed potent antiviral activity, and dramatically reduced the infectivity of a SARS-CoV-2 pseudovirus within minutes (Figure 1). Specifically, AcT-PDA composite coatings at 10 µg∙mL−1 AcT, 5 mM PGD, and 2.5 mM H2O2 were capable of eliminating >80% of functional viral titer for a lentivirus (using HEK293T as susceptible target cells) and >70% of functional viral titer for a SARS-CoV-2 pseudovirus (using HEK293T-Ace2 as susceptible target cells) upon exposure to the composites and the substrates in just 5 min, which is consistent with rapid decontamination. Thus, the AcT-PDA composite was effective in neutralizing both VSV-g and SARS-CoV-2 pseudovirus particles in a very short time, which mimics simple surface contact.
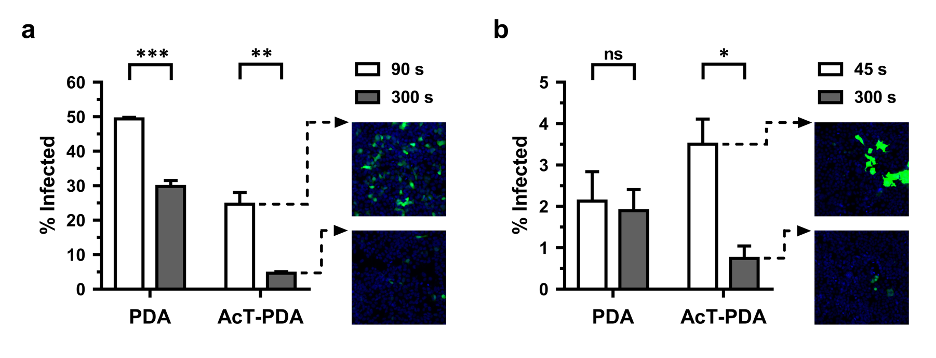
Figure 1. Antiviral activity of AcT (10 μg∙mL−1)-PDA composite against different viruses. (a) VSV-g pseudotyped lentivirus and (b) SARS-CoV-2 pseudovirus in 300 s at 25oC. The x-axis shows the condition tested and the y-axis shows the % infected. The black bars represent t = 90 s and t = 45 s for the lentivirus and pseudovirus, respectively, while the gray bars represent t = 300 s. The representative images show the virus-infected cells after treatment with AcT-PDA at initial time point (t = 90 s for lentivirus and t = 45 s for pseudovirus) and at 300 s. Nuclei are stained blue with Hoechst 33342. A student’s t-test was used to calculate statistical significance. Error bars represent mean ± SEM across three replicates. * p < 0.05, ** p < 0.01, ns-not significant. [Wang et al. Sci. Rep. 11, 12410, 2021]
As one of the most significant public health emergencies in recent decades, the outbreak of COVID-19 raised considerable concern regarding viral pandemics. However, coronaviruses are not the only threats we face, as the occurrences of other infectious viruses, such as monkeypox, respiratory syncytial virus, seasonal influenza, and emerging avian influenza, continue to arise. Extensive research has demonstrated that viral invasion involves not only the interaction of viral surface proteins with host cell receptors but also their interaction with negatively charged glycosaminoglycans (GAGs) on the host cell surface. Heparan sulfate (HS) acts as a facilitator or potentially a co-receptor during SARS-CoV-2 invasion. This intriguing role of HS hints at a promising strategy to disrupt virus-host cell interactions by using sulfated polysaccharides, which can mimic HS, to competitively bind to the virus and block viral infection.
Early into the COVID-19 pandemic, we demonstrated that the well-known anticoagulant heparin, a highly negatively charged homologue of heparan sulfate (HS), exhibits exceptional binding affinity to the spike protein (S-protein) of SARS-CoV-2 [Kim et al., Antiviral Research, 181, 104873 (2020)]. The S-protein of SARS-CoV-2 binds more tightly to immobilized heparin (KD ≈ 10⁻¹¹ M) than the S-proteins of either SARS-CoV (KD ≈ 10⁻⁷ M) or MERS-CoV (KD ≈ 10⁻⁹ M). We further evaluated the in vitro antiviral properties of heparin and other closely related polysaccharides to assess the relevance of heparin-related glycosaminoglycans (GAGs) and other sulfated polysaccharides as part of a pharmacopeia of potential therapeutics targeting SARS-CoV-2 [Kwon et al., Cell Discovery, 6, 50 (2020)]. Specifically, heparin, HS, other GAGs, and the marine sulfated polysaccharide, fucoidan, were screened using surface plasmon resonance (SPR) to measure their binding affinities to the SARS-CoV-2 S-protein (Figures 2 and 3). Our results reveal that specific sulfated polysaccharides, including heparin, TriS-heparin, and fucoidan, bind tightly to the S-protein of SARS-CoV-2 in vitro, suggesting that they can act as decoys to interfere with S-protein binding to the heparan sulfate co-receptor on host tissues.
To model this, we constructed a docking model between heparin and the S-protein receptor binding site (RBD) using the crystal structure of the chimeric RBD-ACE2 complex (PDB ID: 6VW1). The RBD’s amino acid residues involved in binding the ACE2 (angiotensin-converting enzyme 2) receptor also participated in heparin binding, suggesting a mechanism of viral entry inhibition by heparin. Antiviral activities correlated with the SPR results. The most potent compound tested, RPI-27, is a high molecular weight, branched polysaccharide related to the known compound fucoidan, and had an EC50 of 8.3 ± 4.6 μg/mL, which corresponds to approx. 83 nM. This is substantially more potent than remdesivir having a reported in vitro EC50 value of 770 nM in Vero-E6 cells and 11.4 µM in Vero-CCL81 cells, currently approved for emergency use for severe COVID-19 infections. The smaller RPI-28 has the same basic structure as RPI-27 but a lower molecular weight and, thus, a lower activity (1.2 μM). Heparin and the TriS-heparin (an intermediate in the bioengineered heparin synthesis pathway) also have potent antiviral activity withEC50 values of approx. 2.1 and 5.0 μM, while the lower molecular weight NACH had an approx. EC50 of 55 μM. Similar antiviral activity of heparin has also been demonstrated recently. Heparin and TriS-heparin are similar, with the latter devoid of the relatively small fraction of 3-O-sulfate groups present on heparin. Thus, their similar activity is expected. However, the low molecular weight NACH had far lower antiviral activity. Less sulfated GAGs, such as heparan sulfate and various chondroitin sulfates, because of their very low S-protein binding were not tested in the antiviral assay.
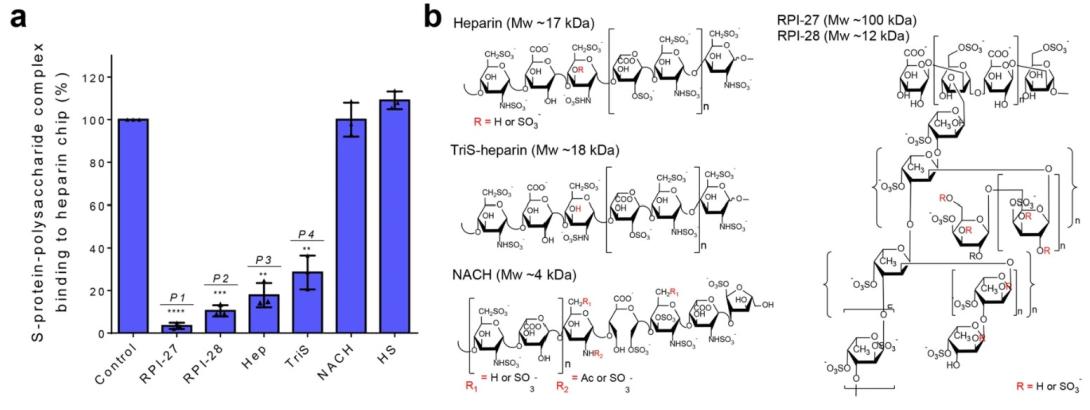
Figure 2. a) Surface plasmon resonance (SPR) experiments were used to screen polysaccharides that outcompete immobilized heparin binding to SARS-CoV-2 S-protein. Data are presented as mean ± s.d., n = 3 biologically independent samples. A two-sided t-test was performed to test significance against the control (P1 < 0.0001, P2 = 0.0003, P3 = 0.0016, P4 = 0.0041). b) Structural units comprising polysaccharides used for in vitro antiviral studies.

Figure 3. c) Focus reduction assay images of virus infection on treatment of indicated polysaccharides. At 48 h after infection, Vero cells were fixed and probed with SARS-CoV-2 spike primary antibody (1:10000, Sino Bio Inc.) and HRP-conjugated goat rabbit (1:10000, Abcam) secondary antibody. d) Vero cells were infected with SARS-CoV2 at a MOI of 2.5 × 10-3 at different doses of each polysaccharide for 48 h. The viral yield was quantified using a focus reduction assay. Cytotoxicity in Vero cells was measured using a WST-1 assay. The left and right y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the polysaccharides, respectively. Cytotoxicity experiments were performed in duplicate with n = 3 biologically independent samples. Focus reduction assay experiments were performed in mean ± s.d. (quadruplicate measurements) with n = 3 biologically independent samples. e) The RBD-ACE2 binding interface is stabilized by an extensive hydrogen bonding network involving sidechains of several residues on both RBD and ACE2. Polar sidechains of N487, Y489, Q493, Q498 and Y505 on the spike protein RBD along with other residues would be able to bind to heparin and inhibit RBD-ACE2 interaction. Heparin (here an octasaccharide) forms a hydrogen bond network with N448, N450, Q493 and N501 that aids in its occupancy of this binding regions and sterically restrict access to Q498, Y489 and Y505 necessary for ACE2 receptor binding. [Kwon et al. Cell Discov. 6, 50 (2020)]
Although some specific sulfated polysaccharides have demonstrated anti-SARS-CoV-2 activity in vitro, their in vivo efficacy requires further validation. We explored several marine sulfated polysaccharides and confirmed their in vivo anti-SARS-CoV-2 activity, indicating potential for prophylaxis [Song et al., Carbohydrate Polymers, 337, 122156 (2024)]. Specifically, we identified and structurally characterized several fucoidan extracts, including those from different species of brown macroalgae, as well as a rhamnan sulfate from a green macroalga species (Figure 4). A high-molecular-weight fucoidan extracted from Saccharina japonica (FSjRPI-27) and a rhamnan sulfate extracted from Monostroma nitidum (RSMn) showed promising competitive inhibition of spike glycoprotein receptor binding to a heparin-coated SPR chip, with IC₅₀ values < 50 ng/mL. This inhibition was also observed in cell-based assays using hACE2 HEK-293T cells infected with pseudotyped SARS-CoV-2 virus, yielding IC₅₀ values < 1 µg/mL. Effectiveness was further demonstrated in vivo using hACE2-transgenic mice. Intranasal administration of FSjRPI-27 provided protection when dosed 6 hours prior to and at the time of infection, and then every two days post-infection, achieving 100% survival with no toxicity at 10⁴ plaque-forming units per mouse compared to buffer controls. At a 5-fold higher viral dose, FSjRPI-27 reduced mortality and lowered viral titers in bronchoalveolar fluid and lung homogenates compared to buffer controls.
Figure 4. Framework of research on the anti-SARS-CoV-2 activity of marine sulfated polysaccharides and their structure-activity relationships.
Sulfated polysaccharides are not limited to activity against coronaviruses. We have also determined that they are effective against monkeypox [Shi et al., Molecules, 27(18), 5898 (2022)] and respiratory syncytial virus (RSV) [Shi et al., Frontiers in Molecular Biosciences, 10, 1151174 (2023)]. With an increasing understanding of structure-activity relationships, we are designing and tailoring sulfated polysaccharides into glycomaterials, pharmaceuticals, and unique adjuvants applicable in the fight against existing and emerging viruses.
The effectiveness of fucoidans in competitive binding and cell-based studies suggested that they could be useful as a prophylactic glycan against SARS-CoV-2 infection. Consistent with the in vitro study, i.n. treatment with S. japonica fucoidan (FSjRPI-27) before and during an LD50 SARS-CoV-2 infection of hACE2 transgenic mice led to significant protection, with 100% survival and no noticeable morbidity, as shown by the absence of weight loss (Figure 5). Treatment with FSjRPI-27 alone in the absence of infection likewise led to no changes in weight, indicating a lack of fucoidan toxicity. In contrast, FSjRPI-28-treated mice showed levels of mortality and weight changes similar to PBS-treated infected mice. We conclude that FSjRPI-27, but not FSjRPI-28, can mediate significant in vivo protection against SARS-CoV-2 infection.
The protective efficacy of FSjRPI-27 was further evaluated using a five-fold greater SARS CoV-2 infection dose. In this case, 85% of PBS-treated mice succumbed to infection with substantial weigh loss (20-35%) following challenge with SARS CoV-2 virus (Figure 6). In contrast, approximately two-thirds of the FSjRPI-27 treated mice survived and regained their body weight starting on day 7 post-infection. At 15 dpi, none of the surviving mice showed any apparent signs of disease.
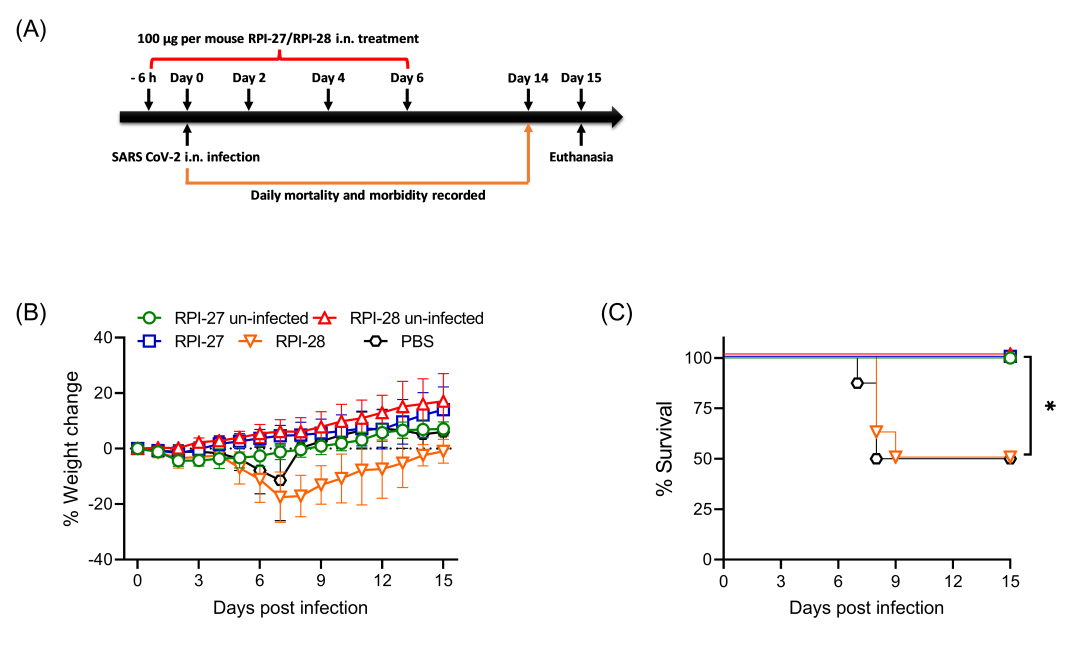
Figure 5. Morbidity and mortality of mice after LD50 pneumonic infection with SARS-CoV-2 virus. A, Groups of mice were inoculated i.n. with 100 µg of FSjRPI-27 or PBS at 6 h before infection, at the time of infection and at 2, 4 and 6 dpi. The mice were challenged i.n. with 1 × 104 PFU/mouse of SARS-CoV-2 purified virus on Day 0. Weight change (B) and survival (C) were monitored for 15 days post infection. Eight mice were used in each group. Survival was analyzed by Log rank Mantel-Cox test. *, P < 0.05.
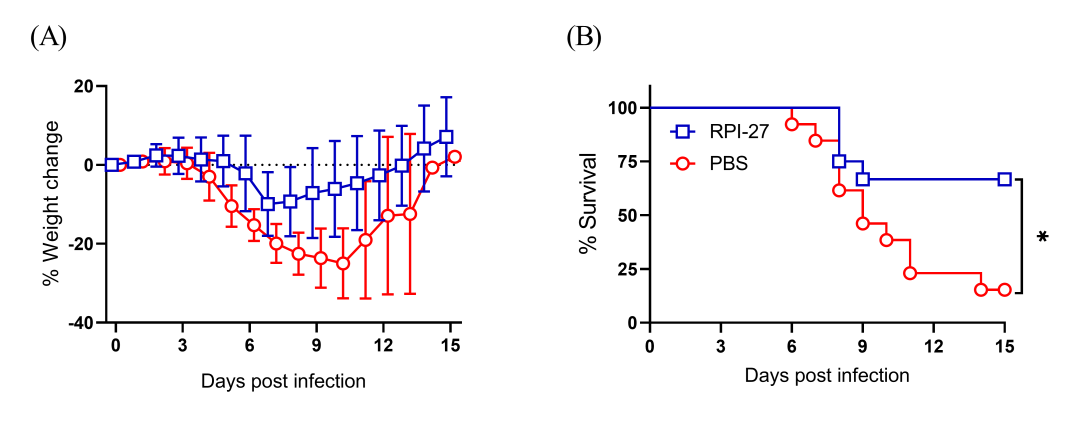
Figure 6. Morbidity and mortality of mice after 5×LD50 pneumonic infection with SARS-CoV-2 virus. Groups of mice were inoculated i.n. with 100 µg of FSjRPI-27 or PBS at 6 h before infection, at the time of infection and at 2, 4 and 6 dpi. The mice were challenged i.n. with 5 × 104 PFU/mouse of SARS-CoV-2 purified virus on Day 0. Weight change (A) and survival (B) were monitored for 15 days post-infection. Twelve mice (6 male and 6 female) in each group were used to assess mortality and morbidity. Survival was analyzed by Log rank Mantel-Cox test. *, P < 0.05.
Current Collaborators:
Fuming Zhang – Rensselaer Polytechnic Institute
Jung Joo Hong – Korea Research Institute of Bioscience and Biotechnology
Dennis Metzger and Amit Singh - Albany Medical Center
