Recovery of recombinant proteins and plasmids from E. coli cytoplasm depends on cell disruption by mechanical, chemical, and/or enzymatic methods, which usually cause incomplete cell breakage or protein denaturation. We have designed controllable autolytic E. coli strains to facilitate purification of recombinant proteins and plasmid DNA that is based on a programmable autolytic E. coli platform that is highly efficient, in which cell lysis is initiated upon induced expression of T4 lysozyme with N-terminal fusion of a cell-penetrating peptide (CPP) [Zha et al. J. Agric. Food Chem. 69, 3134-3143, 2021]. Through engineering of the CPP sequence and copy number, and by incorporating the fusion lytic gene into the E. coli genome, more than 99.97% of cells could be lysed within 30 min of induction regardless of cell age (Figure 1). We further tested the expression and release of a recombinant enzyme in this strain using lysostaphin (Lst) as a model and demonstrated that 4 h induction of the lytic gene after 3 h of Lst expression resulted in 98.97% cell lysis. In addition, Lst obtained from this system had the same yield, yet 1.63-fold higher activity, compared with that obtained from cells lysed by freeze-thawing and sonication. This autolytic platform shows potential for use in large-scale microbial production of proteins and other biopolymers, including plasmids for mRNA vaccine and immunotherapy technologies.
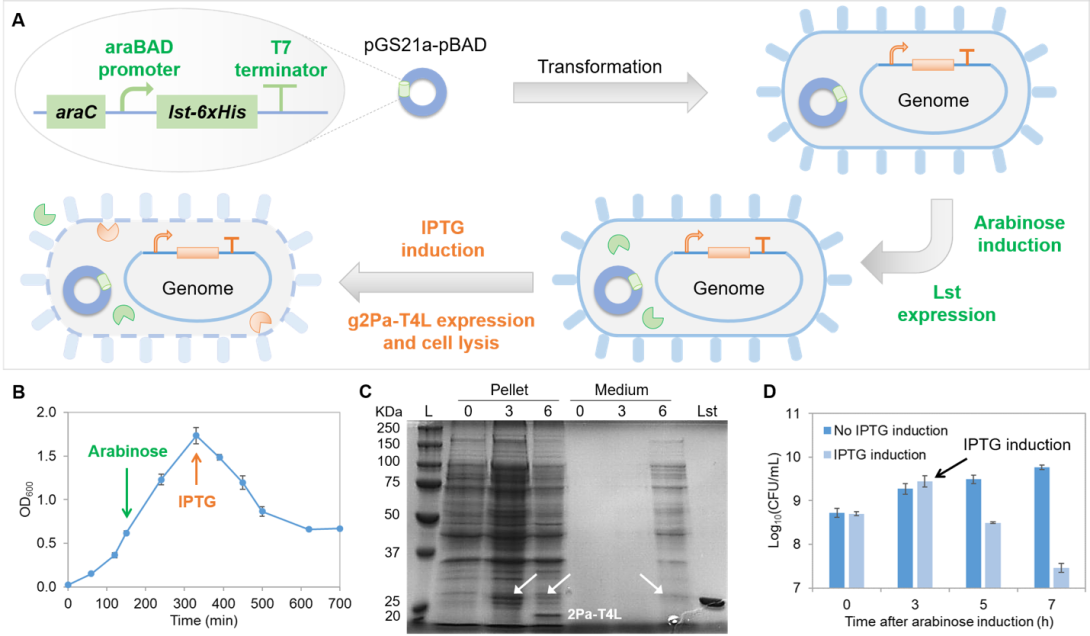
Figure 1. Release of intracellularly expressed Lst in the autolytic E. coli BL21 (DE3) system containing g2Pa-t4l and plasmid pGS21a-pBAD-Lst. (A) Schematic of the induced lysis of E. coli and release of intracellular Lst. (B) Change of culture turbidity with arabinose induction (green arrow) and IPTG induction (orange arrow). (C) Distribution of intracellular proteins in the cell pellet and the culture medium during the course of Lst and g2Pa-T4L expression as analyzed by SDS-PAGE: arrows indicate Lst; 0, 3, 6 indicate time of arabinose addition, 3 h after arabinose addition (Lst expression, time of IPTG addition), and 3 h after IPTG induction; L indicates molecular weight ladder, and the last lane of the gel indicates purified Lst. (D) Change in cell viability with or without IPTG induction after arabinose induction for Lst expression; the time point of arabinose addition is set as zero. [Zha et al. J. Agric. Food Chem. 69, 3134-3143, 2021]
Current Collaborators:
Xia Wu and Jian Zha – Shaanxi University of Science and Technology
