Cell lytic enzymes are essential in targeting Gram-positive bacteria by degrading their peptidoglycan cell walls. These enzymes typically exhibit a two-domain structure: an N-terminal catalytic domain that performs enzymatic cleavage and a cell wall binding domain (CBD) that anchors the enzyme to its bacterial target. This modular architecture enables lytic enzymes to deliver both precise targeting and potent antimicrobial activity, making them powerful tools in combating bacterial infections.
Leveraging the inherent modularity of lytic enzymes, CBDs have proven to be highly versatile, allowing for the discrimination of closely related bacterial species. By isolating and engineering CBDs independently from their catalytic domains, we have developed innovative strategies for pathogen detection or antimicrobial interventions.
CBDs have demonstrated extraordinary specificity when coupled with silver nanoparticles for selective bacterial targeting. We have demonstrated this approach by using CBD-BA, the binding domain from B. anthracis, which selectively bound B. anthracis even in mixtures containing B. subtilis or S. aureus. This coupling converted the broadly antimicrobial silver nanoparticles into a highly selective antimicrobial agent, effectively targeting pathogenic bacteria while minimizing disruption to normal microflora. This biologically-assisted hybrid strategy presents a promising method for selective bacterial decontamination [Kim et al. ACS Appl. Mater. Interfaces 10, 13,317-13,324 (2018)] (Figure 1).
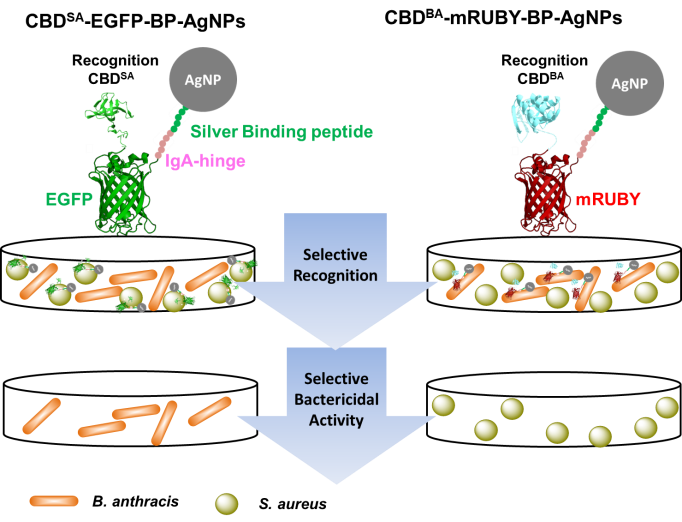
Figure 1. Schematic representation of cell wall binding domain-fluorescent protein-silver binding peptide complexation with AgNPs for selective recognition and specific killing of target pathogenic bacteria; cell wall binding domain CBDSA against S. aureus and CBD-BA against B. anthracis [Kim et al. ACS Appl. Mater. Interfaces 10, 13,317-13,324 (2018)].
The modular nature of cell lytic enzymes can be exploited through unique assemblies (Figure 2). Cell lytic enzymes consist of catalytic and cell wall binding domains, which can be swapped among those of other lytic enzymes to produce unnatural chimeric enzymes. We have shown that microbially-generated biotinylated catalytic domains (CD) and cell wall binding domains (CBD) from the bacteriocin lysostaphin (Lst) and a putative autolysin from Staphylococcus aureus (SA1) can be assembled with streptavidin to form chimeric lytic enzymes [Kim et al. Biomacromolecules 20, 4035-4043, 2019].
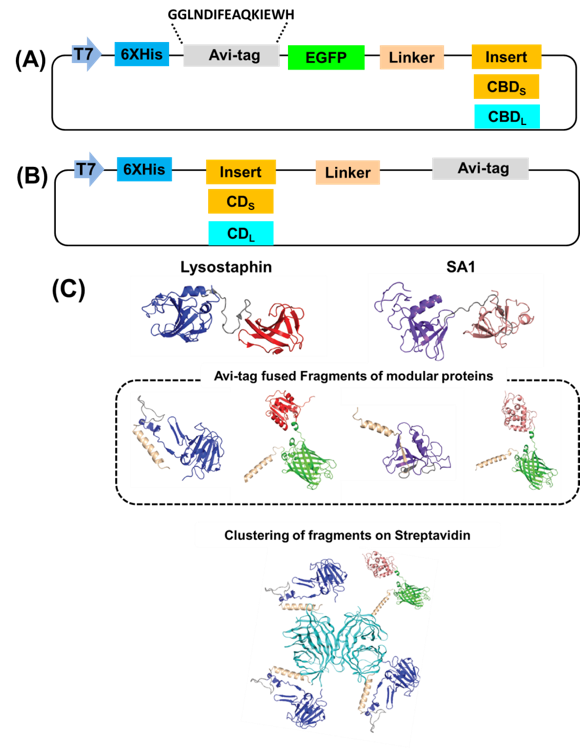
Figure 2. (A) Vector construction for enhanced green fluorescent protein (EGFP)-cell wall binding domain (CBDS) from SA1 and cell wall binding domain (CBDL) from lysostaphin fusions with Avi Tag at the N-terminus. (B) Vector construction for the CDS from SA1 and CDL from lysostaphin with Avi Tag at the C‐terminus. (C) Schematic representation of the modular assembly of recombinant catalytic domains (CDs) and cell wall binding domains (CBDs) with a biotinylated on streptavidin. Blue and red colors indicate catalytic and cell wall binding domains of lysostaphin, respectively, and magenta and brown colors indicate catalytic and cell wall binding domains of SA1, respectively. Gray color indicates linker regions. Cyan color indicates streptavidin.
By self-assembling CBDs with streptavidin and biotinylated reporters, such as glucose oxidase and DNA barcodes, we have developed multiplex detection systems capable of identifying pathogens like Staphylococcus aureus, Bacillus anthracis, and Listeria innocua. These systems integrate ELISA and qPCR methods to achieve detection limits as low as 10 CFU/mL (Figure 3), demonstrating both sensitivity and practicality for applications in clinical diagnostics and environmental monitoring [Kwon et al., Anal. Chim. Acta. 2018, 1030, 156-165].
Figure 3. (A) Schematic illustration for detecting whole bacteria using the CBD-SA-DNA barcode complexes. (B) limit of detections (LODs) of three different bacteria, S. aureus, B. anthracis, and L. innocua cells via the qPCR assay of DNA-barcoded target bacteria [Kwon et al., Anal. Chim. Acta. 2018, 1030, 156-165].
In addition, CBDs have been paired with split fluorescent proteins (FPs) to multimerize both CBDs and signaling molecules to enhance detection sensitivity of target bacteria. Multimeric FP-CBD fusions allow ultra-sensitive detection with limits as low as 0.1 ng for pathogens like S. aureus and B. anthracis. The multimeric design also facilitates simultaneous detection of multiple bacterial species and supports enzymatic signal amplification for even higher sensitivity [Xu et al, New Biotechnol. 2024, 82, 54–64].

Figure 4. Schematic representation of the multimeric split fluorescent protein-fused cell wall binding domains (FP-CBD)n for specific bacterial detection via fluorescence and enzymatic signal generation [Xu et al, New Biotechnol. 2024, 82, 54–64].
