Broad-spectrum antibiotics indiscriminately kill bacteria, removing non-pathogenic microorganisms and leading to evolution of antibiotic resistant strains. Specific antimicrobials that could selectively kill pathogenic bacteria without targeting other bacteria in the natural microbial community or microbiome may be able to address this concern. Cell lytic enzymes are natural antimicrobial agents that depolymerize cell wall peptidoglycans of target bacteria and cause rapid cell lysis [Wu et al. Annu. Rev. Chem. Biomol. Eng. 8, 87-113 (2017)]. There are four types of cell lytic enzymes – endolysins, autolysins, virion-associated lysins (VALs), and bacteriolysins (Figure 1). Endolysins and VALs are bacteriophage-associated lytic enzymes. VALs act externally at the beginning of phage infection for the local decomposition of target cell wall and injection of phage genome into the host cell, whereas endolysins degrade cell wall from inside the host cell at the end of the infection cycle for release of phage progeny. Autolysins and bacteriolysins are generated by bacterial cells. While autolysins function on the producing cells and take part in cell wall remodeling and cell division, bacteriolysins lyse the species that compete for nutrients and growth space, and hence protect the producing cells. These enzymes are exquisitely selective with narrow killing spectra and are environmentally friendly [Bhagwat et al. Appl. Microbiol. Biotechnol. 104, 9019-9040, 2020]. In addition, it is exceptionally difficult for targeted bacteria to gain resistance against lytic enzymes.
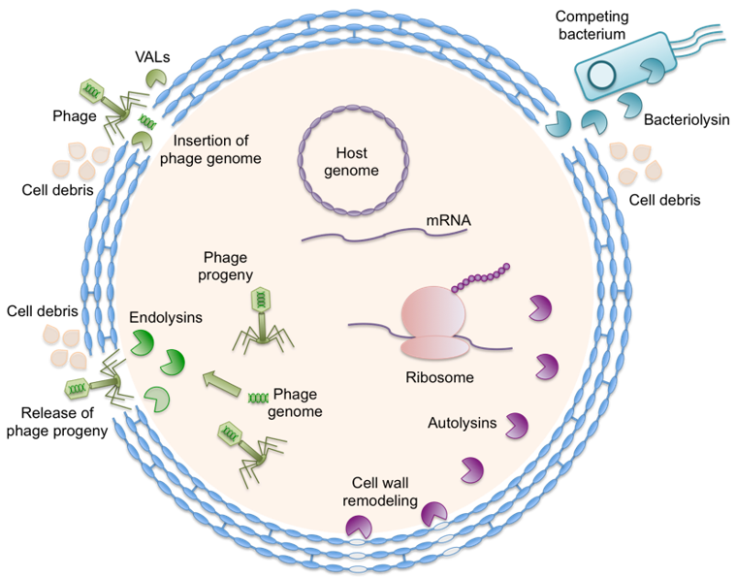
Figure 1. Different types of cell lytic enzymes with actions against a microbial target [Wu et al. Annu. Rev. Chem. Biomol. Eng. 8, 87-113 (2017)].
In particular, there continues to be a need for developing efficient and environmentally friendly treatments for Bacillus anthracis, the causative agent of anthrax. One emerging approach for inactivation of vegetative B. anthracis is the use of bacteriophage endolysins or lytic enzymes encoded by bacterial genomes (autolysins) with highly evolved specificity toward bacterium-specific peptidoglycan cell walls. We performed in silico analysis of the genome of B. anthracis strain Ames, using a consensus binding domain amino acid sequence as a probe, and identified a novel lytic enzyme that we termed AmiBA2446 [Mehta et al. Appl. Environ. Microbiol. 79, 5899-5906 (2013)]. This enzyme exists as a homodimer, as determined by size exclusion studies, and possesses N-acetylmuramoyl-l-alanine amidase activity, as determined from LC-MS analysis of muropeptides released due to the enzymatic digestion of peptidoglycan. Phylogenetic analysis suggested that AmiBA2446 is an autolysin of bacterial origin. We characterized the effects of enzyme concentration and phase of bacterial growth on bactericidal activity and observed close to a 5-log reduction in the viability of cells of B. cereus 4342, a surrogate for B. anthracis. We further tested the bactericidal activity of AmiBA2446 against various Bacillus species and demonstrated significant activity against B. anthracis and B. cereus strains. We also demonstrated activity against B. anthracis spores after pretreatment with germinants. AmiBA2446 enzyme was highly stable in solution, retaining its activity after 4 months of storage at room temperature.
The ability for cell lytic enzymes to target and kill specific bacteria is strongly dependent on the cell wall characteristics. In many Gram-positive bacteria, cell wall teichoic acids present a barrier to exogenously added lytic enzymes. Moreover, some bacteria slowly become resistant to various classes of peptidoglycan hydrolases, for reasons not well studied, in the presence of growth-supporting nutrients, which are prevalent at sites of infection. We have shown that Staphylococcus aureus, a human and animal pathogen, while susceptible to the potent staphylolytic enzyme lysostaphin (Lst) in buffered saline, is highly resistant in the rich medium tryptic soy broth (TSB). Through a series of biochemical analysis, we identified that the resistance was due to prevention of Lst-cell binding mediated by the wall teichoic acids (WTAs) present on the cell surface. Inhibition or deletion of the gene TarO responsible for the first step of WTA biosynthesis greatly reduced S. aureus resistance to Lst in TSB. To overcome the resistance, we took advantage of the gene regulation potential of CRISPR-dCas9 and demonstrated that downregulation of TarO, TarH, and/or TarG gene expression, the latter two encoding enzymes that anchor WTAs in the outer layer of cell wall peptidoglycan, sensitized S. aureus to Lst and enabled eradication of the bacterium in TSB in 24 h (Figure 2). As a result, we elucidated a key mechanism of Lst resistance in metabolically active S. aureus and provide a potential approach for treating life-threatening or hard-to-treat infections caused by Gram-positive pathogens [Wu et al. Biotechnol. Bioeng. 116, 3149-3159, 2019].
Figure 2. Lysostaphin (Lst) is a potent bacteriolytic enzyme against the pathogenic bacterium Staphylococcus aureus. However, in nutrient-rich environment, the bacterial cell surface glycopolymer wall teichoic acid (WTA) blocks the binding of Lst to S. aureus and helps the cells to resist the action of Lst. By using CRISPR-dCas9 targeting key steps in WTA biosynthesis, Lst can interact with and degrade S. aureus cell wall peptidoglycan, thus causing cell lysis and death.
We are discovering and mechanistically characterizing a wide range of lytic enzymes that target key human pathogens. In addition to identifying new enzymes and uncovering enzyme mechanisms, we are exploiting these enzymes for applications in cell lysis for recovery of recombinant proteins and plasmids, infrastructure decontamination and pathogen detection.
