Deciphering and controlling the wealth of intrinsic (e.g., expressed transcription factors) and extrinsic (e.g., chemicals and growth factors) signals involved in the self-renewal and differentiation of stem cells remains a challenge in cell biology that is hampering the use of stem cells in screening and clinical applications. The development of technologies that enable high throughput, high content cell-based screening of agents and culture conditions that impact multiple cellular features is expected to aid in elucidating complex interactions among the multiple signals involved in developmental pathways. This information will deepen our understanding of how stem cells differentiate or self-renew to enhance the control and efficiency of generating stem cell derived cell-based models.
Following previous work with non-human stem cells and somatic cells [Fernandes et al. Biotechnol. Bioeng. 106, 106-118, 2012], we are printing human stem cells in three-dimensional (3D) high-throughput cellular microcultures using a microarray ‘chip’ format wherein the cells can be maintained, grown, induced to differentiate, and analyzed (Figure 1). This is done by embedding the cells in a hydrogel, such as alginate or Matrigel, printing the gel as spots as small as 20 nL and allowing gelation of discrete hemispherical cell spots followed by culture/incubation in media. Notably, a 3D culture provides cells with a microenvironment that can more accurately mimic the geometrical, mechanical, and biochemical aspects of different tissues. Following culture preparation and screening, these whole-cell microarrays may be analyzed to assess endpoints such as protein expression using in-cell, on-chip immunofluorescence, as well as cell growth, viability, and morphology in a high-throughput, high-content manner [Meli et al. Stem Cell Res. 13, 36-47, 2014].
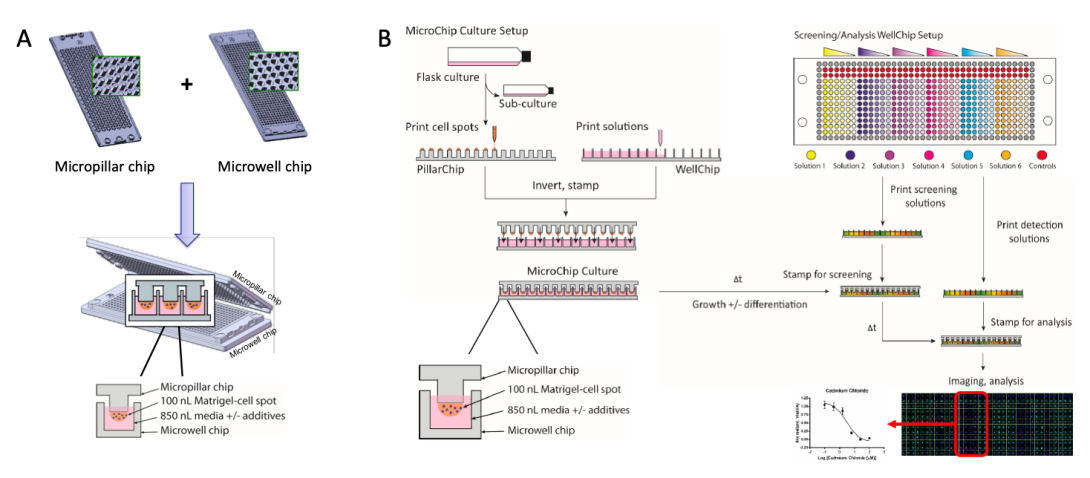
Figure 1. (A) The microarray chip system consists of complementary micropillar (PillarChip) and microwell (WellChip) formats, wherein cell spots are prepared atop pillars on the PillarChip and cultured in WellChips, which contain varied media compositions (e.g., presence of compounds at different concentrations). Each cell spot is cultured in a discrete microwell, which enables up to 532 unique conditions to be screened in parallel per PillarChip. (B) The generic workflow of preparation and screening on-chip cultures using the microarray chip system.
We have begun to assess the influence of matrix and ECM components and other compounds on self-renewal and differentiation of human embryonic stem cells (hESCs) and human neural stem cells (hNSCs) using in-cell, on-chip immunofluorescence analysis of key marker proteins (Figure 2) [Nierode et al. Stem Cell Rep. 7, 5, 970-982 (2016)]. Cultures can be addressed either in groups of a few spots, or individually by means of complementary chips and/or add-on chambers that allow their incubation with components and marker antibodies of interest. Of particular interest is the evaluation of broader signaling pathways involved in self-renewal and differentiation.
This has been extended to identify conditions for guided and specific differentiation of human stem cell and progenitor cells for continued development and engineering of in vitro cell culture systems for use in regenerative medicine, drug discovery, and human toxicology [Nierode et al. Biotechnol. Bioeng. 116, 168-180 (2019]. We proceeded to demonstrate the use of a microarray chip-based platform to screen, in high-throughput, individual and paired effects of 12 soluble factors on the neuronal differentiation of a human neural progenitor cell line (ReNcell VM) encapsulated in microscale 3D Matrigel cultures. Dose-response analysis of selected combinations from the initial combinatorial screen revealed that the combined treatment of all-trans retinoic acid (RA) with the glycogen synthase kinase 3 inhibitor CHIR-99021 (CHIR) enhances neurogenesis while simultaneously decreasing astrocyte differentiation, whereas the combined treatment of brain-derived neurotrophic factor and the small azide neuropathiazol enhances the differentiation into neurons and astrocytes. Subtype specification analysis of RA- and CHIR-differentiated cultures revealed that enhanced neurogenesis was not biased toward a specific neuronal subtype. Together, these results demonstrate a high-throughput screening platform for rapid evaluation of differentiation conditions in a 3D environment, which will aid the development and application of 3D stem cell culture models.
We are also investigating the role of glycans in the hESC environment in early development using immunofluorescent analysis of protein expression coupled with glycan analysis. Finally, the differential responses of NSCs and hESCs to drug targets and toxins as compared to adult, terminally differentiated, primary cells and transformed cell lines are being studied using on-chip viability and toxicity screens [Mikael et al. Stem Cells Devel. 28, 278-289 (2018)].
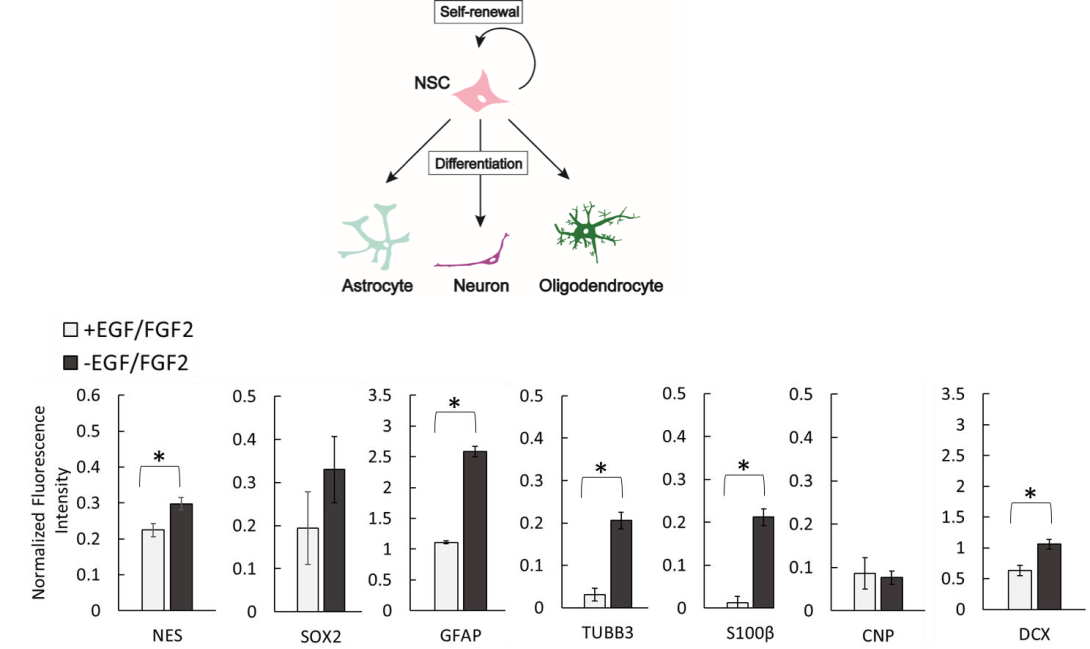
Figure 2. (Top) Human neural progenitor cells (hNPCs) are multipotent cells capable of either self-renewal or differentiation into various neural cell types (including astrocytes, neurons and oligodendrocytes). By assessing the expression levels of key protein markers that are expressed within the specific cell types (e.g. SOX2 in an undifferentiated hNPC or DCX in a differentiating neuron), we can follow the progression of differentiation and characterize cells cultured on-chip in 3D under various conditions. (Bottom) A spontaneous growth factor removal-induced differentiation of an immortalized human neural progenitor cell line (ReNcell VM) was analyzed using in-cell, on-chip immunofluorescence to quantify the extent of a 10-day differentiation in microscale 3D cultures on the chip platform [Nierode et al. Biotechnol. Bioeng. 116, 168-180 (2019].
